Gene Expression Regulation
Research area: Bioinformatics, Computational Chemistry and Systems Biology
WORK IN PROGRESS
Keywords: computational biology, transcriptional regulation, cancer genomics, epigenetics, RNA dynamics, RNA metabolism, RNA modifications, single-molecule sequencing, Nanopore sequencing, nascent RNA
Research interest
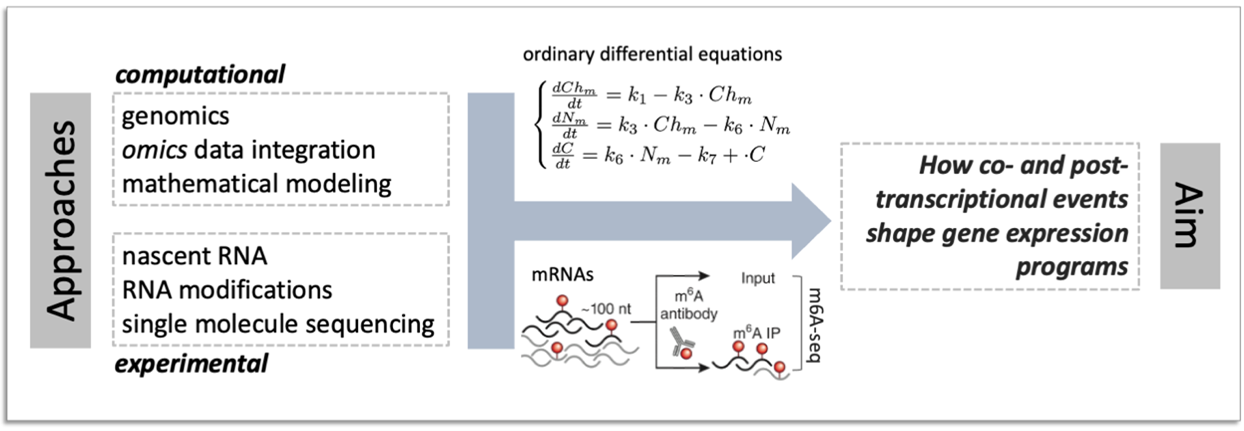
We study the complex life of RNA molecules, which are continuously synthetized, processed, translated and ultimately degraded. We develop methods to decipher how the combined action of machineries responsible for individual steps of the RNA life cycle shapes gene expression programs.
We aim at deciphering the role of RNA modifications (including, while not limited to, the N6-methyladenosine) as determinant of RNA fate.
Ongoing projects in the group are directed at unravelling the role of RNA modifications in cancer, and in particular in the context of: i) gene expression programs driven by the MYC oncogene, ii) resistance to BET inhibitors, iii) spliceosomal vulnerability.
Group members

Mattia Pelizzola - Group Leader
room 3059, building U3-BIOS, tel.: +39 02 6448 3207
lab 4045, building U4, tel. +39 02 6448 3393
mattia.pelizzola@unimib.it
Google Scholar, Linkedin, Twitter (@MattiaPelizzola)
BtBs profile , UNIMIB profile , Publications (Bicocca Open Archive)

Research projects
-2021-present: Principal investigator on a project funded by AIRC (IG2020 ID.24784), focused on the role of RNA modifications in the onset of aberrant gene expression programs in breast cancer.
-2021-present: Supervisor for a 3-years AIRC fellowship to Lucia Coscujuela, focused on the role of RNA modifications in liver cancer drug resistance.
-2022-present: Supervisor for a 3-years AIRC fellowship to Mattia Furlan, focused on the role of RNA modifications in breast cancer spliceosomal vulnerbility.
Selected articles
-Coscujuela Tarrero L, Famà V, D'Andrea G, Maestri S, de Polo A, Biffo S, Furlan M, Pelizzola M. Nanodynamo quantifies subcellular RNA dynamics revealing extensive coupling between steps of the RNA life cycle. Nat. Commun. 15, 7725 (2024). doi: 10.1038/s41467-024-51917-2
-Furlan M, de Pretis S, Pelizzola M. Dynamics of transcriptional and post-transcriptional regulation. Brief Bioinform 22, bbaa389 (2020). https://doi.org/10.1093/bib/bbaa389
-Furlan M, Galeota E, Del Gaudio N, Dassi E, Caselle M, de Pretis S, Pelizzola M. Genome-wide dynamics of RNA synthesis, processing, and degradation without RNA metabolic labeling. Genome Res 30, 1492–1507 (2020). DOI: 10.1101/gr.260984.120
-de Pretis S, Kress TR, Morelli MJ, Sabò A, Locarno C, Verrecchia A, Doni M, Campaner S, Amati, B, Pelizzola M. Integrative analysis of RNA polymerase II and transcriptional dynamics upon MYC activation. Genome Res 27, 1658–1664 (2017). DOI: 10.1101/gr.226035.117.
-Sabò A*, Kress TR*, Pelizzola M*, de Pretis S, Gorski MM, Tesi A, Morelli MJ, Bora P, Doni M, Verrecchia A, Tonelli C, Fagà G, Bianchi V, Ronchi A, Low D, Muller H, Guccione E, Campaner S, Amati B. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature 511, 488–492 (2014). https://doi.org/10.1038/nature13537
Pelizzola’s Lab – #PelizzolaLab_BtBs
Dipartimento di Biotecnologie e Bioscienze (BtBs) - Università degli Studi di Milano-Bicocca, Milano, Italia
last update: April 2025